Suspicions of two bridgehead invasions of Xylella fastidiosa subsp. multiplex in France – Communications Biology
A low polymorphism within ST6 and ST7 genome sequences but enough temporal signal to date divergence time
A set of 82 genome sequences of X. fastidiosa subsp. multiplex composed of all publicly available ones, as well as newly acquired sequences were analyzed in this study (Supplementary Data1). They represented diverse geographical origins (Brazil n=1, France n=52, Italy n=3, Spain n=12, USA n=14), spanned over 36 years of evolution (19832018), and were centered on the most frequent lineages detected in France, i.e., ST6 and ST7. Most genome sequences (85.4%) were Illumina dye sequences. Average genome length was 2,520,537bp, with a mean N50 and L50 of 354,785bp and 7.28, respectively. The length of their core genome alignment was 1,679,574bp, in which 16,739 single nucleotide polymorphisms (SNPs) were detected. After removal of regions with evidence of recombination 13,818 SNPs only originating from mutation events were kept.
Considering ST6 and ST7 strains genome clustering from all geographic origins using FastSTRUCTURE on non-recombinant SNPs, the patterns find was in partial discordance with traditional MLST grouping (Fig.S1). While all French and American ST6 strains clustered in a homogeneous cluster, as expected from MLST, Spanish ST6 strains were scattered within the ST7 cluster grouping all the French ST7 strains, the three American ST7 strains, and one of the Spanish ST81 strain. The two other Spanish ST81 strains were intermediate between French ST7 and the Spanish ST6 strains. American, Italian and Brazilian X. fastidiosa subsp. multiplex strains of other STs grouped into three other clusters (Fig.S1). Similar groupings were observed by plotting the core genome SNPs from mutation on a heatmap (Fig.S2). The low polymorphism observed in the core genome and the absence of sub-structuration within French ST6 or ST7 genome sequences were not compatible with any further population genetics analysis aiming at reconstructing the pathways of the emergence of X. fastidiosa at a regional scale in French territories based only on these data.
In contrast, the complete set of 82 genome sequences, isolated all over the world, was proved suitable for investigation of the timing of X. fastidiosa subsp. multiplex divergence (Supplementary Data1). Analyzing the linear regression of sample age against root-to-tip distance (Fig.S3) and performing a date randomization test (DRT) with BEAST36 (Fig.S4) revealed a sufficient temporal signal at the whole-tree scale. Therefore, the dataset could be used to coestimate evolutionary rates with ancestral divergence times with a tip-based calibration approach.
At the scale of the genome, the mean substitution rate for X. fastidiosa subsp. multiplex was inferred at 3.2165107 substitutions per site per year (95% Highest Posterior Density [HPD] 1.54681075.2043107 substitutions per site per year). The standard deviation of the uncorrelated log-normal relaxed clock calculated by BEAST was 1.020, suggesting moderate variation in rates among branches. The divergence between French and American ST6 strains was estimated in 1987 (19741994 95% HPD) (Fig.1). Then the divergence between these strains and the Spanish ST81 strains was estimated in 1425 (7661914 95% HPD). The one between ST7 French and American strains was estimated in 1971 (19241994 95% HPD). This time to the most recent common ancestor (TMRCA) was due to the USA RAAR6Butte strain, as the TMRCA with the two other ST7 USA strains (M12 and Griffin-1) is older (=1510; 9911911 95% HPD). The split between Spanish ST6 strains and ST7 strains was estimated in 1027 (1201743 95% HPD). The divergence between the group of ST7 and Spanish ST6 strains and the group of ST81 and ST6 French and American strains was estimated in 755 (397 to 1540 95% HPD).
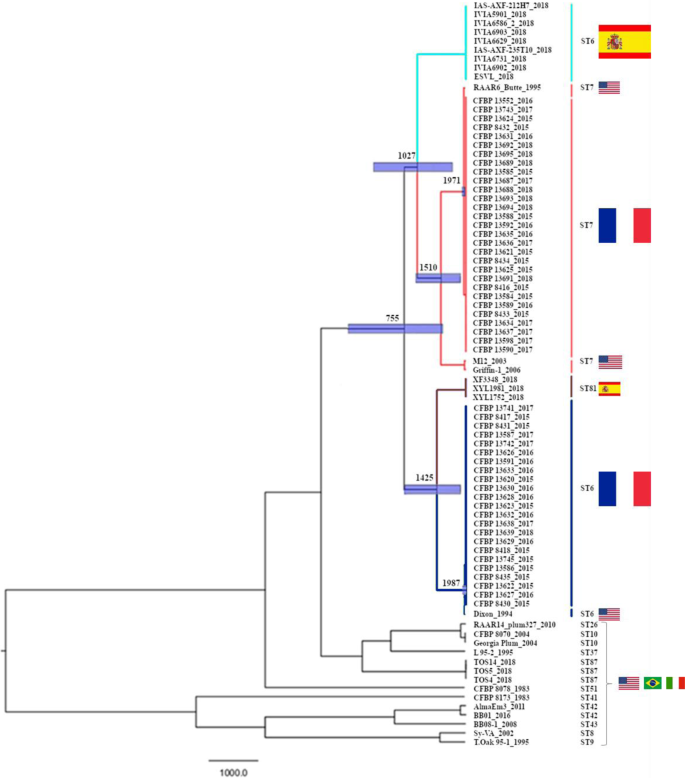
Genealogy was inferred by Maximum-likelihood phylogenetic inference using BEAST 2.6.1. and a GTR model, based on SNPs variations among the 82 genome sequences. For details on the data, refer to Supplementary Data1. The tree main sequence types are highlighted in blue (ST6), red (ST7) and brown (ST81) and flags refer to the country of isolation of the strains. Node bars refer to 95% HPD.
The development of a MLVA efficient on DNA extracted from plant samples revealed that French X. fastidiosa split into four groups of strains
To elucidate the scenario that led to the establishment of X. fastidiosa subsp. multiplex in Corsica and PACA, variable number of tandem repeats (VNTRs) were used to complement the low information gained from SNP calling. An in silico analysis of the X. fastidiosa subsp. multiplex strain M12 genome sequence was performed to search for new VNTRs, in order to complete the set of available ones37,38. A set of 13 VNTRs was selected and while initially developed to discriminate strains of the subspecies multiplex, it proved to be valid for use on all other X. fastidiosa subspecies (Table1, Supplementary Data2, Fig.S5). The developed VNTR-13 scheme was then optimized for direct use on DNA extracted from plant samples, due to the difficulty of isolating the strains and to make use of the large amount of infected plant samples available in France. The complete setup of the VNTR-13 scheme including its validation is detailed as supplementary results.
In France, from 2015 to 2018, among the plant samples tested in the framework of the French surveillance and officially declared to be X. fastidiosa infected, 917 samples had a Cq<32 (the amplification limit Cq of the MLVA), which corresponded to ~5105 X. fastidiosa cells per gram of plant. Depending on the availability of frozen samples and in order to avoid the analysis of several samples from the same foci, a selection of samples was made to maximize the number of foci and plant species analyzed and resulted in a set of 534 samples for the MLVA. A total of 396 samples were successfully genotyped for all 13 loci (184 ST6 and 212 ST7; i.e. 43.18% of the French X. fastidiosa samples available and 74.16% of the tested samples) (Table2, Supplementary Data3). The loci were all highly polymorphic across the French dataset with a number of alleles ranging from 7 to 21 and a number of TRs ranging from 1 to 30 (Table3). Simpsons diversity index ranged from 0.52 to 0.88 and allelic richness from 3.82 to 15.73. For the VNTR loci ASSR-19, XFSSR-40 and XFSSR-58, all possible allele sizes of the allelic range were observed within the collection. For the VNTR loci ASSR-9, ASSR-11, ASSR-12, ASSR-16, COSS-1, GSSR-7, OSSR-16, OSSR-19, XFSSR-37 the observed diversity of TR sizes did not cover the entire allelic range as one to three TR sizes were not observed, indicating either missing infected samples in the evolution path or large mutation steps (Supplementary Data3). MLVA accurately resolved the different haplotypes from the French outbreaks as more than 95% of the haplotypes were detected with 12 markers (Fig.S6). The discriminatory power of the MLVA was 0.9969, showing a very high level of discrimination.
For a minority of 13 French X. fastidiosa infected plant samples, several peaks were observed on the electrophoregrams on 3 up to 12 amplified loci, and this was confirmed in at least two independent analyses. As some alleles were, for now, specific of ST6 or ST7 (e.g., for ASSR-16:29 TR=ST6, 24 TR=ST7), these results revealed the presence of co-infections by several strains in these plants and for some of them potentially by several STs.
MLVA allowed observing a large haplotype diversity within French ST6 and ST7 X. fastidiosa as 320 haplotypes were delineated, among the 396 samples (Supplementary Data3). The 184 ST6 samples were genotyped in 148 haplotypes, while the 212 ST7 samples were genotyped in 172 other haplotypes, and no VNTR profile was shared between these two STs (Supplementary Data3 and 4, Fig.S7). The distribution of allele frequencies for each of the 13 VNTR loci did not indicate differences between samples isolated in Corsica or PACA or their host plant. ST6 samples were grouped into four clonal complexes (i.e., networks grouping haplotypes differing from their closest neighbor at a single VNTR locus). The founder haplotype (#309) of the main clonal complex grouped 11 samples from three plant species (P. myrtifolia, Lavandula x allardii, Calicotome villosa), and was linked, in this clonal complex, to 95 samples that were all sampled in Corsica (Fig.2). ST7 samples grouped into 15 clonal complexes. The founder haplotype (#138) of the main clonal complex comprised 15 samples of three plant species (P. myrtifolia, Genista x spachiana, Helichrysum italicum) and was linked to 88 samples, of which 86 were isolated in Corsica and only two in PACA. The other 17 smaller clonal complexes grouped from two to eight samples that were sampled in a same region, with the exception of one clonal complex that grouped haplotype #163 sampled in PACA and haplotype #165 sampled in Corsica. The remaining 167 samples represent singletons, differing by at least two loci (=80 ST6 haplotypes and 83 ST7 haplotypes) or by three and more loci (=36 ST6 haplotypes and 32 ST7 haplotypes) (Supplementary Data3).
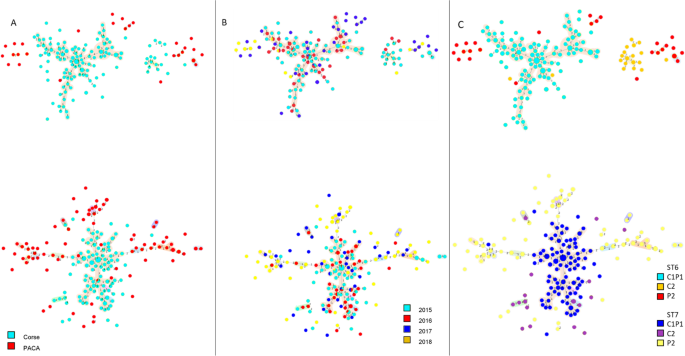
Dot diameter represents the number of strains per haplotype. Link number refer to the number of TR loci polymorphic and distinguishing two haplotypes. Dot colors refer to A sampling region; B year of sampling; C ABC grouping. Colored area around haplotypes the clonal complexes.
Due to the nature of the data that mostly came from official monitoring programs, which aim is the eradication of any infected plant and not population genetics studies, it was impossible to analyze the impact of the host and year of sampling on the minimum spanning tree (MST) structure. Nevertheless, the presence of infected samples from 2015 in distal parts of evolutionary branches and of founder haplotypes sampled in 2017 (Fig.2) suggests that sampling was carried out in an already diversified population. Regarding host plants, a large majority (69.95%) of our dataset was made of myrtle-leaf milkwort plants (P. myrtifolia), while Spanish broom (Spartium junceum) accounted for 7.07% of the samples and then all remaining 23 plant species each accounted for less than3.78% of the samples, which was highly imbalanced and did not allow the analysis of MST structuration relative to host species. Moreover, 4.68% of the haplotypes were identified in more than one plant species, indicating that haplotypes did not face intrinsic dispersal barriers.
In order to retrace the routes of dissemination, populations that can be analyzed by Bayesian methods were sought. On the basis of genetic clustering analyses (DAPC and STRUCTURE) and geographical information, we validated the clustering of all 396 samples into four clusters that were, as expected, also supported (p-value<0.05) by RST and FST pairwise comparisons (Fig.3, Fig.S8, Supplementary Data5, See supplementary results for complete description of the clusters).
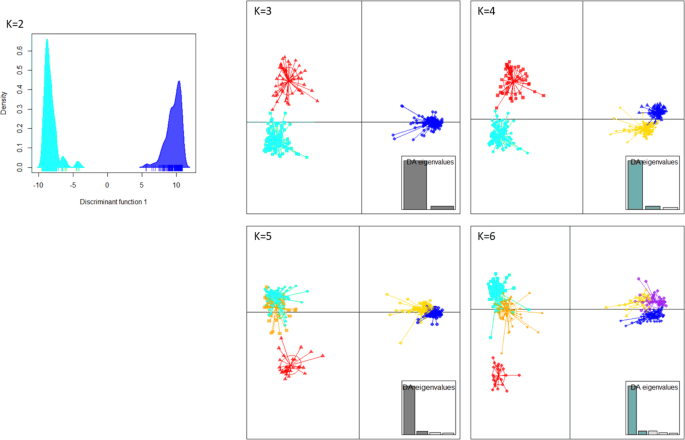
The eigenvalues showed that the genetic structure was captured by the first two principal components retained onto axis 1 (horizontal axis) and axis 2 (vertical axis). The dots represent the individuals, and the clusters are shown as inertia ellipses. Clusters turquoise, red and orange grouped ST6 samples and clusters blue, yellow, and purple grouped ST7 samples.
Bridgehead introductions of X. fastidiosa subsp. multiplex ST6 and ST7 in Corsica from PACA
We inferred the routes of dissemination of X. fastidiosa subsp. multiplex in France using Approximate Bayesian Computation, beginning with the definition of a set of evolutionary scenarios that may explain the observed data. Firstly, in order to keep tested scenarios as simple as possible, we analyzed ST6 and ST7 French samples separately, because there was no known element indicating that these different strains were introduced simultaneously into France and tip-dating provided different dates of divergence for French ST6 and ST7 strains from their American relatives. Secondly, we had to define groups of strains likely to have shared common history (in sexual species, these would be populations, but in bacteria this step is not trivial). In each cluster previously defined by DAPC at k=4, samples originating from distinct regions were split as subgroups when the corresponding populations were differentiated based on RST and FST pairwise comparisons, with a p-value<0.05 (Supplementary Data3 and 5). Moreover, analysis of molecular variance evaluated that the majority of the genetic variance occurred within the subgroups (ST6: 63.99%, ST7: 78.12%) (Supplementary Data6). As a result, three groups were defined for each ST (i.e. one American and two French, Supplementary Data3, Fig.4). For ST6, the DAPC cluster 1 included all samples of the main clonal complex plus a few singletons. This group of samples was mainly isolated from Corsica (135 samples) and a few from PACA (two samples) and was named ST6_C1P1. The DAPC cluster 2 was composed of all the other singletons and two of the small clonal complexes grouping two haplotypes. It was separated into two subgroups: one named ST6_C2, grouping the 19 samples originating from Corsica and another one named ST6_P2 grouping the 28 samples originating from PACA. For ST7, the DAPC cluster 3 included all samples of the main clonal complex plus a few singletons. This group of samples were mainly isolated from Corsica (110 samples) and a few from PACA (eight samples) and was named ST7_C1P1. The DAPC cluster 4 was composed of all the other singletons and 13 of the small clonal complexes of two haplotypes. The DAPC cluster 4 was separated into two subgroups, one, named ST7_C2, composed of the 19 samples originating from Corsica, and another one, named ST7_P2, composed of the 75 samples originating from PACA.
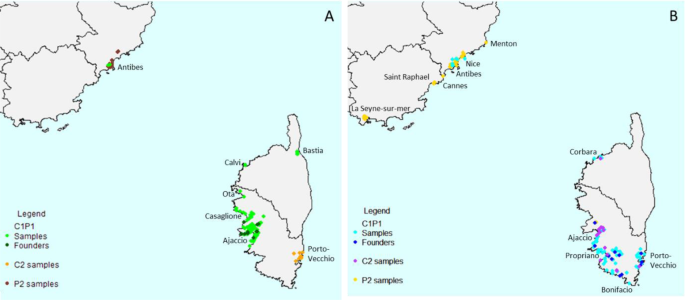
A ST6 plant samples and strains, B ST7 plant samples and strains.
The number of scenarios with three French populations, one outgroup population and possible non-sampled (ghost) populations is huge, making it impossible to perform a single analysis to answer our question. To cope with this complexity, we adopted a two-step approach composed of (i) a bottom-up step in which populations from a same ST were analyzed two by two in three different analyses, which aimed at deciphering histories between pairs of French populations (Fig.S9A), and (ii) a top-down step confronting three-population scenarios not excluded by the bottom-up step (Fig.S9B).
On the three French ST6 populations (C1P1, C2, and P2), after bottom-up approach, no scenario (topology combination of populations) achieved sufficiently high posterior probability and low prior error rate to be considered as the best scenario to reconstruct the evolutionary history of the ST6 French populations in regards to their USA counterparts. However, some scenarios had so low posterior probabilities (p-value<0.05) that they could be ruled out (Supplementary Data7). Specifically, scenarios 2 and 6 testing the possibility of only one French population were ruled out for all combinations of populations, confirming that the French ST6 samples did group into three populations (Fig.S9A).
Scenarios not excluded during this first step were used in the top-down approach to elaborate scenarios of the evolutionary history of the three French ST6 populations (Fig.S9B). Considering all combinations of populations, 30 scenarios were confronted (Fig.S10). Scenario II.7 was selected in the abcrf analysis as the most probable scenario with 9.9% of the votes and the highest posterior probability (0.260.37) (Supplementary Data7). In this scenario, ST6 strains would have been first introduced in PACA (ST6_P2 population). Then population ST6_C1P1 would have diverged from this initial focus, and would thus represent the first established population in Corsica. A second independent population (ST6_C2) would have diverged later on from the first established ST6_P2 population in PACA, and established in Corsica (Fig.S10). However, it should be tempered by the fact that this selected scenario also presented a high prior error rate (0.72) (Supplementary Data7).
The following most probable scenarios received a low number of votes. The second and third best scenario were III.I.16 and II.9 with respectively 5.8% and 5.7% of votes (Supplementary Data7). These two scenarios had a close topology to scenario II.7, as both considered an introduction in PACA (ST6_P2), from which the population ST6_C2 from Corsica would have diverged. Moreover, on the 30 scenarios confronted, the seven scenarios testing the hypothesis that the Corsican population ST6_C2 would have diverged from the ST6_P2 population from PACA totalized 35.9% of the votes, giving more strength to this event.
Similarly, on the three French ST7 populations (C1P1, C2, and P2), the bottom-up approach did not allow to identified the best scenario, but allowed ruling out scenarios 2 and 6, thereby confirming that the French ST7 samples did group into three populations (Fig.S9A, Supplementary Data7). In addition, scenarios 1 and 9 that tested independent and successive introductions of each French populations from the American ancestor were also excluded. It is however interesting to note that similar scenarios (5 and 10) including an unsampled population between the American ancestor and the French ones were not ruled out. In contrast, scenarios 11 and 12 that were slight variations of previous scenarios 1 and 9 including unsampled populations between the American ancestor and each French populations were ruled out. Altogether, six scenarios (no. 3, 4, 5, 7, 8 and 10, Fig.S9A) were kept for further consideration after this bottom-up approach.
Moving to the top-down approach, based on the results of the bottom-up approach scenarios that tested independent introductions (class I in Fig.S9B) and those testing two independent introductions, one of which was responsible for a bridgehead invasion of the third population (class III.I and III.II in Fig.S9B), were consequently not considered. This left us with two closely related topologies to be tested (Fig.S9B and Fig.S11), i.e., scenarios testing the introduction in France of one population subsequently responsible for two independent bridgehead invasions, (class II in FigS10B), and scenarios considering that the first introduction lead to two successive bridgehead invasions (class IV in FigS10B). Considering all combinations of populations, 12 scenarios were confronted. Scenario II.7 was selected in the abcrf analysis as the most likely scenario with 17.5% of the votes and with the highest posterior probability (0.410.50) (Supplementary Data7). In this scenario, ST7 strains would have been first introduced in PACA (ST7_P2 population). Then population ST7_C1P1 would have diverged from this initial focus, and would thus represent the first established population in Corsica. A second independent population (ST7_C2) would have diverged later on from the first established ST7_P2 population in PACA, and established in Corsica (Fig.S11). However, it should be tempered by the fact that this well supported scenario also presented a high prior error rate (0.700.71) (Supplementary Data7).
Then scenarios having the highest number of votes were scenario IV.27 and scenario IV.25, to which respectively 12.4% and 10.9% of the votes were attributed (Supplementary Data7). Both had a close topology to the best scenario, as they considered that a first population would have been introduced in PACA (ST7_P2), from which populations ST7_C1P1 and ST7_C2 would have diverged but from two successive bridgehead invasions (ST7_C1P1 then ST7_C2 or ST7_C2 then ST7_C1P1) instead of two independent divergence events. Moreover, among the 12 scenarios confronted, the four scenarios testing the hypothesis that the population from PACA (ST7_P2) was the first one introduced in France and from which the other two population diverged totalized 53.24% of the votes, giving more strength to this event.